University of Wisconsin Unified Workflow (UWUW)
The University of Wisconsin Unified Workflow (UW2) aims to solve the issue of running the same Monte Carlo problem using mutiple physics codes. Currently, if you wish to run the same problem in multiple codes you must fully recreate the input file for each code, or maybe write a full syntax translator. UW2 allows users to tag or associate groups of volumes or surfaces with a simple human readable syntax that is translated and stored in the geometry file of a DAGMC problem.
The workflow uses the Python for Nuclear Engineering toolkit, PyNE. We leverage the existing infrastructure in PyNE to allow a consistent transport problem to be defined across all MC codes.
Materials
Materials are the most painful and error-prone items to transfer from code to code, since each MC code specifies materials in a different way. Instead, we tag groups of volumes with a name and syntax that corresponds to material compositions in a predefined material library.
The group naming syntax for describing materials in Cubit is:
CUBIT> group "mat:<Name of Material>"
If you wish to specify a material that is present in the library with a different density, then
CUBIT> group "mat:<Name of Material>/rho:<density>"
So for example, to specify a Stainless Steel at a density of 12.0 g/cc,
CUBIT> group "mat:Stainless Steel/rho:12.0"
Scoring
Each MC code implements tallies, or scores, in very specific ways such that there is sometimes no equivalent to a tally you may be familiar with, code to code. However, there is a Cubit syntax to allow you to request scores on geometric elments. The generic form is
CUBIT> group "tally:ParticleName/ScoreType" add vol x
A specific example, scoring the neutron flux in vol 2,
CUBIT> group "tally:Neutron/Flux" add vol 2
or the photon current crossing surface 3,
CUBIT> group "tally:Photon/Current" add surface 3
Using the underlying PyNE libraries, we can write out the appropriate MC code tally specification snippet; this allows the number of codes that DAGMC supports to grow organically with those that PyNE supports. When PyNE cannot fulfill your tally request it will warn you.
Boundary conditions
All MC codes have an understanding of boundary conditions, and all at least implement some form of graveyard, black hole or region where particles are "killed". Some MC codes go further and define reflecting or other boundary conditions. Furthermore, some apply these boundary conditions to entire volumes as opposed to surfaces. Currently we support only reflecting surface definitions, but in the near term we hope to support full volume reflecting boundaries. One can implement reflecting surfaces in UW2 by adding surfaces to the following Cubit group defintion
CUBIT> group "boundary:Reflecting"
or if you prefer, Lambert (white reflection)
CUBIT> group "boundary:White"
Particle importances
Particle importances are in important aspect of Monte Carlo simulations and are used to help particles to penetrate to "important" regions of the geometry. There are several automatic methods to generate mesh based importances or weights, but if your importances are tied to the geometry, then they can be tagged onto the geometry.
The UW2 workflow has a code agnostic way of defining importances.
CUBIT> group "importance:Neutron/1.0"
This is translated to the code specific version at runtime. Note: Fluka's importance range runs from 1e-5 to 1e5, when written to file. The range is rescaled and any out-of-range values are truncated to 1e-5 or 1e5.
UW2 data
The UW2 data is incorporated into the geometry file (*.h5m) file using the
C++ program uwuw_preproc, the purpose of which is to take the user's
material library, e.g. my_nuc_library.h5, and extract the materials
requested, placing them into the geometry file. If you have already marked up
your geometry using the methods mentioned in previous sections, you can run the
preprocessor.
$ uwuw_preproc <dagmc h5m filename> -v -l <path to nuclear data library>
Be sure to examine the output of this script which will inform you of the materials and densities requested and also the list of tallies that were produced. A sample output is shown below.
$ uwuw_preproc test_geom.h5m -l \
$HOME/.local/lib/python2.7/site-packages/pyne/nuc_data.h5
Also, the program will produce a fatal error if the material is not found in the material library. There is a simulate mode (flag -s) to perform all operations that would be performed on a normal operation, except that no data is actually written. This can be done to test your file to make sure all the expected materials are present in the file.
MCNP5/6-specific steps
The DAG-MCNP input file
Note that all the information here applies to both DAG-MCNP5 and DAG-MCNP6.
The DAG-MCNP input file contains only the data cards section of a standard MCNP input file. There are no cell or surface cards included in the input file.
A new data card has been added to DAG-MCNP to define parameters for the DAGMC geometry capability.
Form: dagmc keyword1=value keyword2=value
check_src_cell: behavior of CEL variable in SDEF card
on [default] standard interpretation for
CEL variable: source rejection
off no cell rejection - assume that
sampled position is in cell CEL
overlap_thickness: allows particle tracking through small overlaps
{real} [default=0.0]
distlimit: toggle usage of flight distance sampled from
physics to accelerate ray-tracing search
off [default] do not use physics flight distance
on do use physics flight distance
Running DAG-MCNP
Running DAG-MCNP is identical to running the standard MCNP, but a few new keywords have been added to the command line to specify the necessary files.
Input Option |
Description |
gcad=<geom_file> |
(required): Specify the filename of the input geometry file.
format (this is the format produced by |
fcad=<facet_file> |
(optional): [default: fcad] Specify the filename of the output facet file. This is the file produced by DAGMC that contains the
geometry as well as the products of a number of
preprocessing steps, which can be quite time-consuming.
This file can be used as input with the |
lcad=<log_file> |
(optional): [default: lcad] Specify the filename of the output log file. This is a skeleton of an MCNP file which contains information about the volumes, surfaces, materials and material assignments, boundary conditions, and tallies defined in the geometry. If you specify a name other than the default for this file on the command-line, that file will be used instead of the one generated automatically by DAG-MCNP. This can be useful if you want to make small changes to your material assignments, importances, etc., but it cannot be used to change anything about the geometry. It is up to the user to ensure that the geometry file and log file being used correspond to each other. This runtime parameter is unique to DAG-MCNP. |
To run DAG-MCNP, you must make the minimum input deck that defines the particle source definition, runtime parameters, and physics cutoffs. You then run DAG-MCNP5 with
$ mcnp5 i=input g=geom.h5m
If DAG-MCNP is unable to find some nuclides, you can modify the lcad file that was produced in the previous step to remove extraneous nuclides, and re-run with
$ mcnp5 i=input g=geom.h5m l=lcad_modified
FluDAG-specific steps
To run a FluDAG-based UWUW problem, like the above MCNP example, the user must
make a minmal Fluka input deck defining runtime parameters and source
definition, remembering to set the GEOBEGIN to FLUGG. Once this is done,
run the mainfludag executable to produce the mat.inp file which contains
all the detailed material assignments and compound descriptions.
$ mainfludag geom.h5m
The user should then paste the contents of mat.inp into the main FLUKA input
deck. Now the user must make a symbolic link to the geometry file named
dagmc.h5m.
$ ln -s geom.h5m dagmc.h5m
The mainfludag executable always looks for the dagmc.h5m file. You can
now run as if it were a standard FLUKA problem
$ $FLUPRO/flutil/rfluka -N0 -M5 -e mainfludag input.inp
OpenMC-specific steps
To run a OpenMC UWUW simulation, a .h5m file containing the UWUW model
must be present in the OpenMC run directory and a dagmc_universe present in
the geometry.xml like so:
<dagmc_universe> filename="dagmc.h5m" id="1" />
OpenMC will then load the geometry and material library when setting up the simulation. For more information on how to use DAGMC geometries in OpenMC, see here.
Geant4-specific steps
To run a DAG-Geant4-based UWUW problem, like those shown above, the user must write a Geant4 macro file that contains at minimum the source description (GPS) and the number of particles to simulate. The problem is then run with
$ DagGeant geom.h5m input.mac
Worked example
Open Cubit, and let's place some volumes to create our first problem. We will create 4 cubes of side 10 cm, shifting each in a different direction.
CUBIT> brick x 10
CUBIT> move Volume 1 x 20 include_merged
CUBIT> group "mat:Lead" add volume 1
CUBIT> group "tally:Photon/Flux" add volume 1
CUBIT> brick x 10
CUBIT> move Volume 2 x -20 include_merged
CUBIT> group "mat:Lead" add volume 2
CUBIT> group "tally:Photon/Flux" add volume 2
CUBIT> brick x 10
CUBIT> move Volume 3 y -20 include_merged
CUBIT> group "mat:Lead/rho:12.3" add volume 3
CUBIT> group "tally:Photon/Flux" add volume 3
CUBIT> brick x 10
CUBIT> move Volume 4 y 20 include_merged
CUBIT> group "mat:Lead/rho:12.3" add volume 4
CUBIT> group "tally:Photon/Flux" add volume 4
CUBIT> brick x 100
CUBIT> brick x 105
CUBIT> subtract volume 5 from volume 6
CUBIT> group "mat:Graveyard" add volume 7
CUBIT> imprint body all
CUBIT> merge all
CUBIT> set attribute on
CUBIT> export acis "example.sat" overwrite
You will end up with something like this.
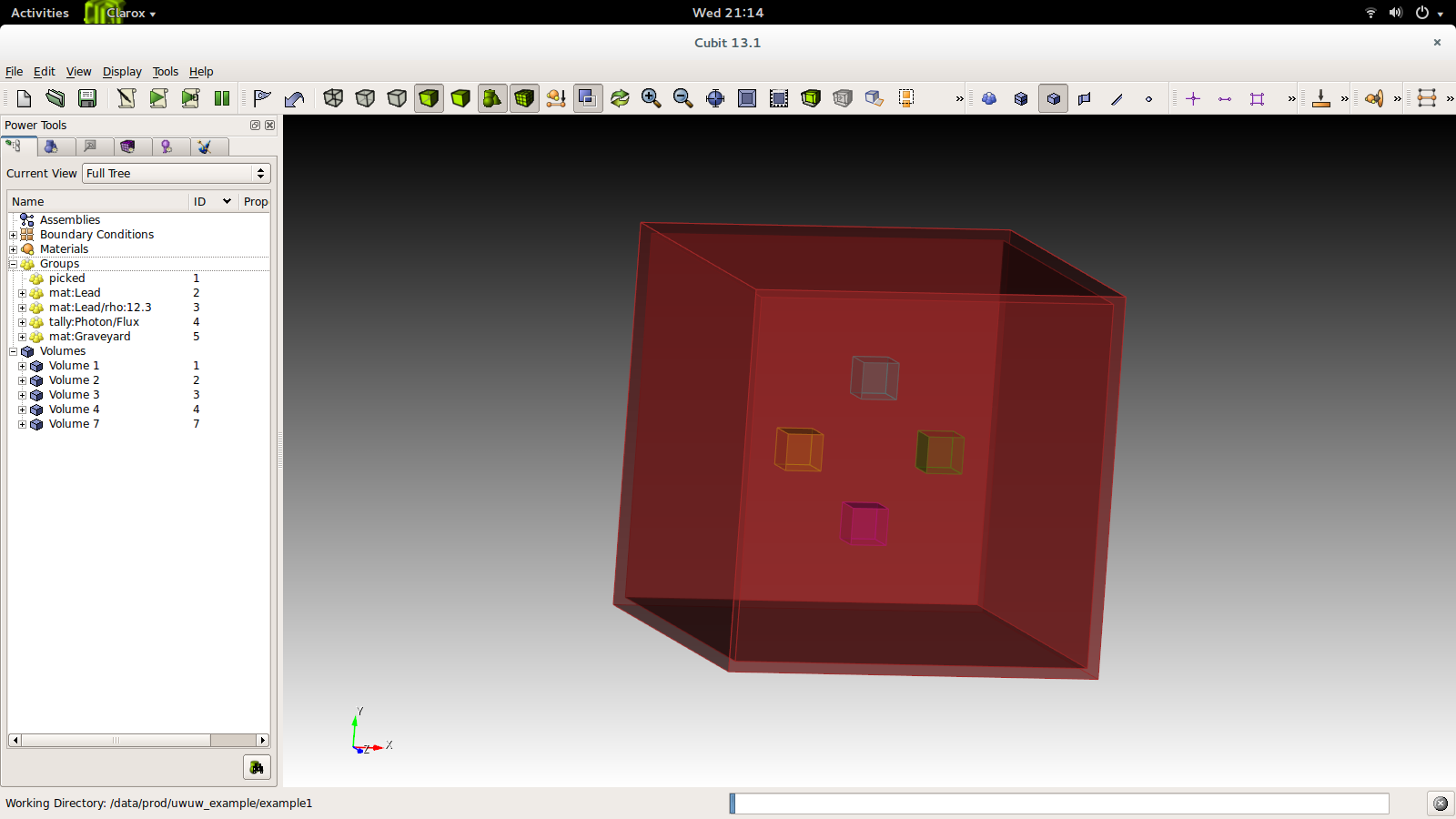
The file is now ready for preprocessing. First we must facet the file:
$ dagmc_preproc example.sat -o example.h5m
Now we can insert all the material data we need:
$ uwuw_preproc example.h5m -l \
$HOME/.local/lib/python2.7/site-packages/pyne/nuc_data.h5
Your output from this step should look exactly the same as this:
Making new material with name : mat:Lead
with fluka_name: LEAD
Making new material with name : mat:Lead/rho:12.3
with fluka_name: LEAD1
Photon PHFL1 3
Photon PHFL2 3
Photon PHFL3 3
Photon PHFL4 3
writing material, mat:Leadwriting material, LEAD to file example.h5m
writing material, mat:Lead/rho:12.3writing material, LEAD1 to file example.h5m
Writing tally PHFL1 to file example.h5m
Writing tally PHFL2 to file example.h5m
Writing tally PHFL3 to file example.h5m
Writing tally PHFL4 to file example.h5m
So we see echoed back to us that we requested a graveyard and two different material assignments: one for lead, as defined in the material library, and another kind of lead at a different density than the library version. We also see that 4 tallies were requested: the photon flux in each volume.
Example input
We are now ready to run DAGMC, once we have made the input deck for each Monte Carlo code. We wish to simulate 10^5 photons with energy 1 MeV emitted isotropically from the point (0,0,0). The inputs for MCNP5 and FLUKA are shown below.
MCNP5 example: let us call this mcnp.inp.
example of UWUW
c notice no cell cards
c notice no surface cards
c notice no blank lines!
sdef x=0.0 y=0.0 z=0.0 par=2 erg=1.0
c notice no materials
c notice no tallies
mode p
nps 1e5
print
FLUKA example: let us called this fluka.inp.
TITLE
* Set the defaults for precision simulations
DEFAULTS PRECISIO
* Define the beam characteristics
BEAM -0.001 10000.0 PHOTON
* Define the beam position
BEAMPOS 0. 0. 0.
* Notice the FLUGG section
GEOBEGIN FLUGG
GEOEND
* notice no material assignments
* notice no scoring assignments
* ..+....1....+....2....+....3....+....4....+....5....+....6....+....7...
RANDOMIZ 1.0
* Set the number of primary histories to be simulated in the run
EMF
START 1.E5
STOP
DAG-MCNP5 run
Now we are ready to run the first DAG-MCNP5 example.
$ mcnp5 i=mcnp.inp g=example.h5m
You should see something like this on screen.
The implicit complement's total surface area = 128550
This problem is using DAGMC version 1.000 w/ DagMC r 0
Using default writer WriteHDF5 for file fcad
/mnt/data/prod/uwuw_example/web_example/example.h5m
Materials present in the h5m file
mat:Lead
mat:Lead/rho:12.3
Tallies present in the h5m file
PHFLUX1
PHFLUX2
PHFLUX3
PHFLUX4
Going to write an lcad file = lcad
Tallies
Thread Name & Version = MCNP5, 1.60
Copyright LANS/LANL/DOE - see output file
_
._ _ _ ._ ._ |_
| | | (_ | | |_) _)
|
comment. photon importances have been set equal to 1.
comment. using random number generator 1, initial seed = 19073486328125
Turned OFF ray firing on full CAD model.
Set overlap thickness = 0
imcn is done
warning. material 1 has been set to a conductor.
warning. material 2 has been set to a conductor.
ctm = 0.00 nrn = 0
dump 1 on file runtpe nps = 0 coll = 0
xact is done
cp0 = 0.01
run terminated when 100000 particle histories were done.
ctm = 0.05 nrn = 900033
dump 2 on file runtpe nps = 100000 coll = 56221
mcrun is done
Feel free to examine the output of the run, but this is only a simple example of what to expect.
FluDAG run
For FluDAG, first we produce the mat.inp snippet file. This must then be
pasted into the full FLUKA input deck.
$ mainfludag example.h5m
The mat.inp file should look like
*...+....1....+....2....+....3....+....4....+....5....+....6....+....7...
ASSIGNMA LEAD1 1.
ASSIGNMA LEAD1 2.
ASSIGNMA LEAD2 3.
ASSIGNMA LEAD2 4.
ASSIGNMA BLCKHOLE 5.
ASSIGNMA VACUUM 6.
*...+....1....+....2....+....3....+....4....+....5....+....6....+....7...
MATERIAL 82. 207.217 11.35 26. LEAD1
MATERIAL 82. 207.217 12.3 27. LEAD2
*...+....1....+....2....+....3....+....4....+....5....+....6....+....7...
* UW**2 tallies
* PHFLUX1
USRTRACK 1.0 PHOTON -21 1.1.0000e+03 1000.PHFLUX1
USRTRACK 10.E1 1.E-3 &
* PHFLUX2
USRTRACK 1.0 PHOTON -21 2.1.0000e+03 1000.PHFLUX2
USRTRACK 10.E1 1.E-3 &
* PHFLUX3
USRTRACK 1.0 PHOTON -21 3.1.0000e+03 1000.PHFLUX3
USRTRACK 10.E1 1.E-3 &
* PHFLUX4
USRTRACK 1.0 PHOTON -21 4.1.0000e+03 1000.PHFLUX4
USRTRACK 10.E1 1.E-3 &
At this point you will need to add two lines to the input file manually. This is because the component of the code which identifies neutron cross section data is not yet complete.
*...+....1....+....2....+....3....+....4....+....5....+....6....+....7....+....
LOW-MAT LEAD1 82. -2. 296. LEAD
LOW-MAT LEAD2 82. -2. 296. LEAD
The lines above must be pasted into the FLUKA input and then run as you would
a native FLUKA problem, with the exception that we give the rfluka script an
executable argument and a new -d argument, which specifies the geometry
filename.
$ $FLUPRO/flutil/rfluka -N0 -M1 -e mainfludag -d example.h5m fluka.inp
The code should run and successfully produce screen output similar to the following. (The filepaths will change according to your system, as will the numerical part of "fluka_26362".)
$TARGET_MACHINE = Linux
$FLUPRO = /mnt/data/opt/fluka/fluka/
Initial seed already existing
Running fluka in /mnt/data/prod/uwuw_example/web_example/fluka_26362
======================= Running FLUKA for cycle # 1 =======================
Removing links
Removing temporary files
Saving output and random number seed
Saving additional files generated
Moving fort.21 to /mnt/data/prod/uwuw_example/web_example/fluka001_fort.21
End of FLUKA run
Dag-Geant4 run
Dag-Geant4 is probably the most trivial of all the UW2-enabled codes to run.
Copy the vis.mac file from DAGMC/geant4/app/vis.mac.
$ DagGeant4 example.h5m
After some loading you should see a GUI window open (if you built Geant4 with visualization enabled). You can then use the Geant4 general particle source to emulate the behaviour of the previous two codes.
Idle> /gps/particle gamma
Idle> /gps/ang/type iso
Idle> /gps/energy 1.0 MeV
Now we are ready to run.
Idle> /run/beamOn 1000000